Mean and standard deviation
This can apply to a population being measured for taxonomic purposes, or observations concerning various individuals such as the results of an experiment where individuals are exposed to different treatments, e.g. drugs.
Once a number of measurements have been taken, it is obvious that the information needs to be converted into a form that simplifies data handling. Usually this involves taking a
mean - often called the average. If all the readings are close together, you can assume that this value is reliable, although in Biology we expect to find more variation than an engineer would expect from a pack of nuts and bolts.
Additionally there are a number of statistics that can be assigned to the data:
If the data collected appear to be normally distributed, the
standard deviation of the mean -
σ - can be used to express the
spread of data about the mean.
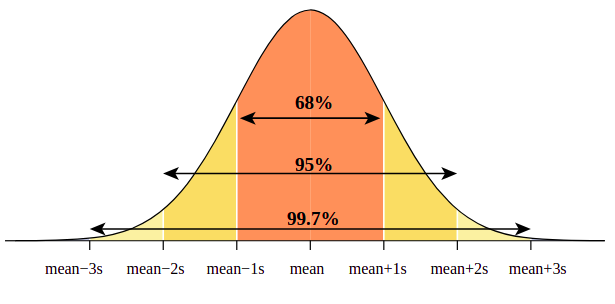
Confidence intervals: The 68- 95 -99.7 rule
68% of data are within 1 standard deviation either side of the mean
95% of data are within 2 standard deviations either side of the mean
99.7% of data are within 3 standard deviations either side of the mean
The
standard deviation of a sample is calculated by summing the
squares of the deviation of each data value from the mean - the difference between each value and the mean - then dividing this by the number of values -1, then taking the square root.
σ = √ Σ ( x - m )2
n-1
where
x = (a series of) values
m = mean of those values
n = number of values in the sample
Population standard deviation: If all the members of a population are measured, the calculation above is divided by n, not n-1. If using a scientific calculator, note that these often have different keys for
σ and
σn-1.
Each value contributes to this statistic.
It effectively reduces the effect of individual anomalies/outliers.
If a number of readings have been taken and used to give points on a graph or column chart, it is normal to use bars (sometimes called error bars) above and below the mean.
Differences between means can be said to be
significant if SDs do not overlap.
In fact, the more standard deviations overlap, the less likely it is that differences are real or significant. In other words they are more likely to be caused by chance.
Sometimes error bars are given as ± 2 standard deviations. These cover 95% of the constituent data points, but this is not a probability measure in itself.
Other sampling statistics
The
standard error of the mean (SEM) is the standard deviation of the mean as calculated above, then divided by the square root of the number in the sample. This measures how far the sample mean of the data is likely to be from the true mean of the population.
SEM = σ
/
√ n
The
range, equal to the top value - bottom value, is less useful as it may overly emphasise (one or two) outlying values, and
nothing in between.
Investigation of genetic diversity in different breeds of dog
One hundred autosomal microsatellite markers distributed across the canine genome were used to examine variation within 28 breeds of dogs (in the USA).
Part of these data relates to different types of
terriers (originally bred to hunt foxes, by digging up their earths or pursuing them down the tunnels leading into the earths), and retrievers (bred to track down and bring back game that has been shot in the field).
| Dog breed |
Mean genetic
diversity
|
S.D. |
| Airedale terrier |
0.515 |
0.020 |
| Jack Russell terrier |
0.758 |
0.012 |
| Yorkshire terrier |
0.684 |
0.018 |
| Labrador retriever |
0.641 |
0.016 |
| Golden retriever |
0.657 |
0.016 |
What do these data show about the differences in genetic diversity between these breeds of dog?
>
Terriers are all different
>
Genetic diversities of Jack Russell > Yorkshire > Airedale
>
Retrievers' diversities are closer/closest
>
S.D.s do not overlap
>
But retrievers' S.D.s are close - and show a (minimal!) overlap)
Using a spreadsheet to calculate standard deviation
Spreadsheet data and display as a simple graphic
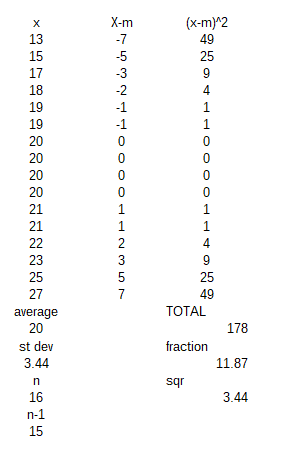
I invented some data to the right to use as an example.
I came upon some data about the mass of a species of mouse and thought I could generate the mean and standard deviation from a list of completely arbitrary numbers.
I have simply assumed that my own mouse data will cover a range of masses from 13g to 27g, with a mean value of 20g.
You will see I have only used integers: whole numbers, and put them in as an ordered list - not as would happen in real life!
The first column shows that highlighting the data then selecting statistical function : STDEV displays the 'standard deviation based on a sample'.
I also set up the calculation step by step on the next 2 columns.
Luckily this gives the same results!
'Standard' scientific calculators can also be used to perform this calculation.
You will probably need to refer to the instruction manual!


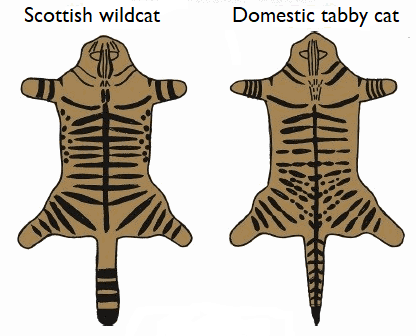 There is no need to kill and skin the animals to identify them!
There is no need to kill and skin the animals to identify them!